Most of the time you will load the dataframe from an external file, most likely a text file, an excel file or a CSV file (Comma-Separated Values). In that case, you may follow this procedure. In brief, you have to use the command read.xls(), read.table() or read.csv() to open a file and transfer its content into R, then store it as an object (dataframe).
One of the usual ways to use the command is the following:
[code language=”r”]
object.name <- read.table("path", header=TRUE, sep=";")
[/code]
where:
object.name is the name of your dataframe in R (choose the name wisely since you’ll have to use it over and over again),path is the whole path to your file, starting with the letter of the drive, unless you have set up the name of the work directory prior to importing the file,; is used to separate the data elements (if not, exchange the comma for the appropriate symbol whenever needed),NB: if you use file.choose() instead of writing a path for the file, R will open an explorer windows and ask you to choose the file from any directory available on your machine. Convenient… In this case, the code is:
[code language=”r”]
object.name <- read.table(file.choose(), header=TRUE, sep=";")
[/code]
An additional step when importing the dataframe is to “attach” it with the command attach(). This step allows you to directly refer to the name of your variables in the commands. Simply type attach(object.name). You may then use name(object.name) to print the name of the variables which you have made accessible with attach(). However, attaching dataframes is often considered as a dangerous move as your R environment might contain vector names matching some of the variables of your dataframe, in which case you end up using vectors and objects which are completely unrelated to your work. See this post for more info. If you do not attach your dataframe, you may refer to the variables in your dataframe using a $ symbol such as in object.name$variable.name. Here is an example (in this example, values were separated by a semi-colon, hence the changed symbol in the sep argument):
[code language=”r”]
my.imported.data <- read.table("d:/dropbox/bioCEED/bioSTATS/CSV/mydata.csv", header=TRUE, sep=";")
attach(my.imported.data)
names(my.imported.data)
[/code]
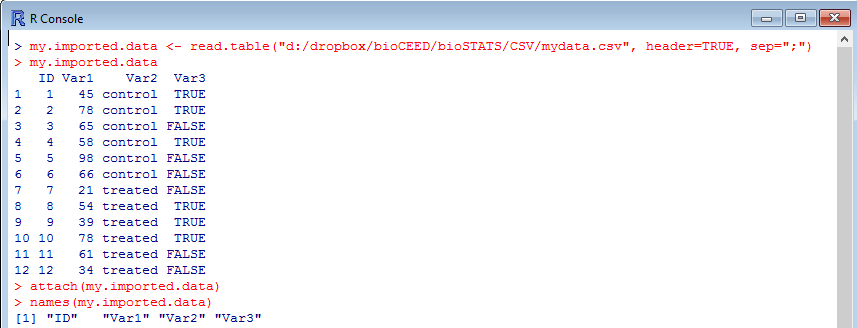
As you see above, the dataframe is well arranged, and everything appears clearly in columns. The variables are revealed by names() and you can clearly count four of them. If you make the mistake not to use the correct symbol as separator, you may see something like this:
[code language=”r”]
my.imported.data <- read.table("d:/dropbox/bioCEED/bioSTATS/CSV/mydata.csv", header=TRUE, sep=",")
attach(my.imported.data)
names(my.imported.data)
[/code]
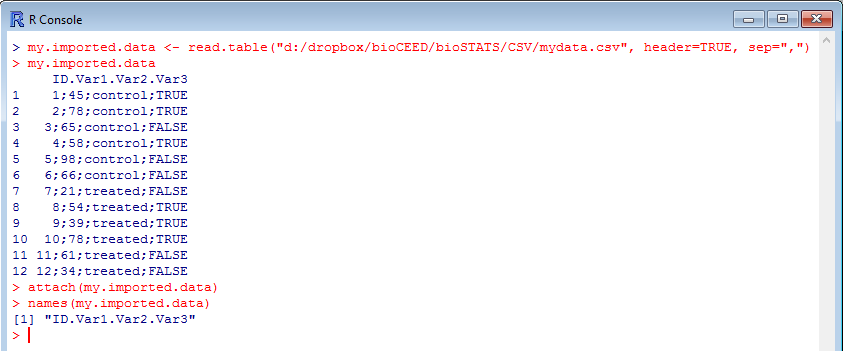
One easily realizes that there is a problem as the table isn’t well aligned, semi-colons are present between the data elements, and the command names() shows only one variable name (which is in fact the concatenation of all 4 variable names). Time to correct the command lines…
