Working with large data sets may be time-consuming or demanding in terms of computer resources due to high number of variables combined with high numbers of observations. Moreover, displaying a table/data frame with many variables in R/Rstudio isn’t very practical and you may soon want to reduce the data set to the exact set of the variables you actually need to work with. A simple, but obvious example of unpractical data sets is BCI in the package vegan. Let’s use it as an example to study the following two dplyr functions that will help you extract variables:
select(): extract a set of given columns/variables from a data frame, by name or by positionpull(): extract the content of a columns/variables and returns them as a vector
For each of these two functions, there is a list of arguments (also called “helpers”) which will help you find precisely the (groups of) variables that you want:
contains(): find variables which name contains the given string of charactersmatches(): find variables which name matches a regular expression or regexstarts_with(): find variables which name starts with a given string of charactersends_with(): find variables which name ends with a given string of charactersone_of(): find variables which name is listed in a given vectorlast_col(): find the last variable
Finally, you may use - in front of each of the above mentioned arguments to drop the variables described by the argument. For instance, while select(last_col()) will ONLY select the last column, select(-last_col()) will select ALL the columns BUT the last one.
Before we go further, you may find more info about BCI HERE. The top of the data frame is rather large due to the 225 (!) labels of the variables; it is in fact so large that it does not display like a table anymore, but rather looks like this:
head(BCI)
Selecting a set of variables with select()
In its simplest form (with no additional argument), select() will extract variables by their full name or by their position in the data frame, while select(-) will drop the variables and return the rest of the data frame. Use commas to list the names and positions, and use colons to define ranges of columns:
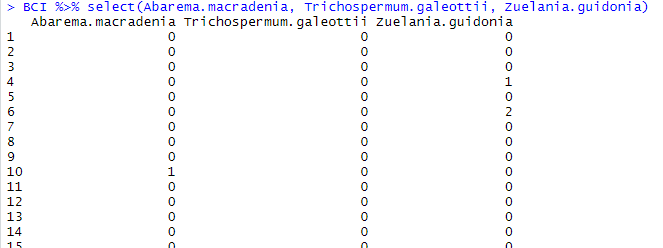
BCI %>% select(Abarema.macradenia, Trichospermum.galeottii, Zuelania.guidonia)
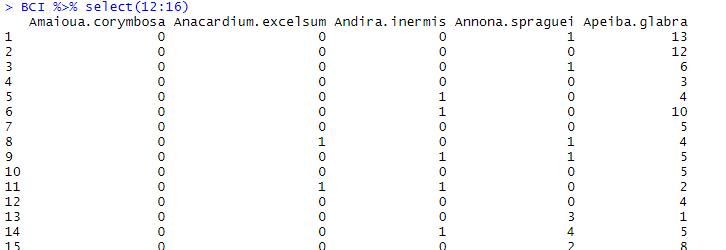
BCI %>% select(12:16)
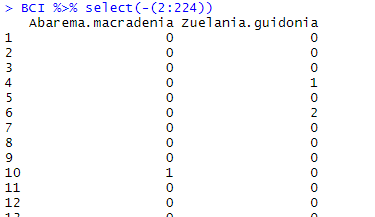
BCI %>% select(-(2:224))
Selecting the content of a variable and returning it as a vector with pull()
BCI %>% pull(56)
BCI %>% pull(Brosimum.alicastrum)
Of course, when extracting something to a vector, it is best to directly set the name of the vector. In this case we would write:
vector.Brosimum.alicastrum <- BCI %>% pull(Brosimum.alicastrum) vector.Brosimum.alicastrum
Useful arguments (“helpers”)
Finding variables which name contains the given string of characters with contains()
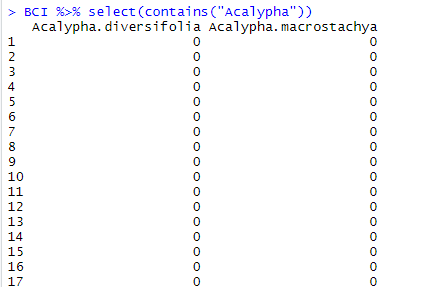
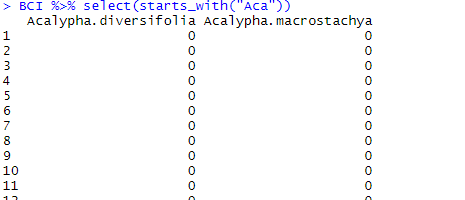
contains() allows for selecting all variables which name contains the string of characters given between parentheses. This might be a fragment of the name or a word. Here in BCI, we want to make sure that we will retrieve all the species of the genus “Acalypha”. We thus write the following code:
BCI %>% select(contains("Acalypha"))
Finding variables which name matches a regular expression (regex) with matches()
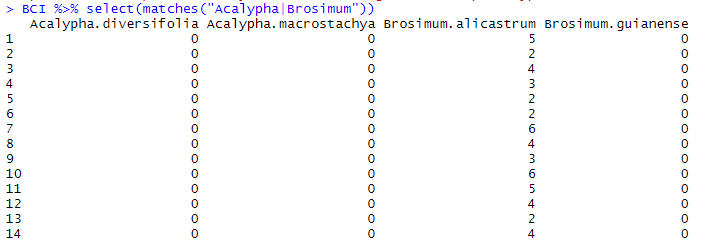
matches() allows for using regular expressions (or regex) to find variables based on patterns or multiple strings. Regular expression are strings that include symbols which replace predefined characters, thus allowing you to pick up patterns in a text or table (for example, a period would replace any character, meaning that a.a would cover all possibilities from aaa to aza), or symbols/operators that mediate a function (for example | means OR, and a|b would mean a OR b). Check here for more info and the list of symbols to use. For example, if we want to retrieve from BCI all the columns that contain EITHER Acalypha OR Brosimum, we will use the following code:
BCI %>% select(matches("Acalypha|Brosimum"))
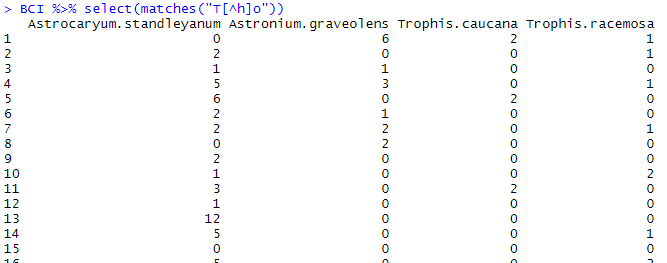
If we want to retrieve from BCI all the columns which contain t.o where . is any character BUT h (and thus avoid picking Zanthoxylum species), we will use the following code:
BCI %>% select(matches("T[^h]o"))
Finding variables which name starts with a given string of characters with starts_with()
starts_with() finds the variables which name starts with the string that you have indicated, logically.
BCI %>% select(starts_with("Aca"))
Finding variables which name ends with a given string of characters with ends_with()
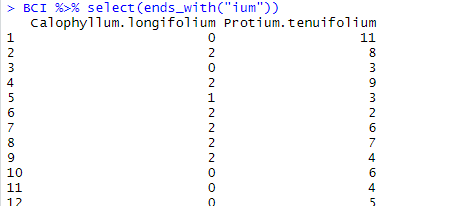
Again, logically, ends_with() finds the variables which name ends with the string that you have indicated.
BCI %>% select(ends_with("ium"))
Finding (all the) variables which name is listed in a given vector with one_of()
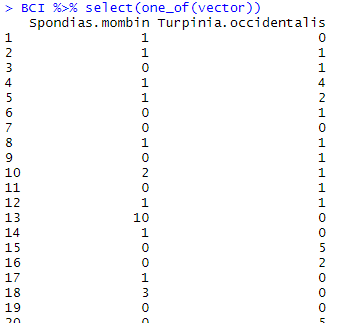
one_of() retrieves all the variables (not only one of the variables) which name is stored in a given vector. Let’s store two names found in BCI in the vector called vector:
vector <- c("Spondias.mombin", "Turpinia.occidentalis")
vector
Let’s now use one_of() to retrieve variables:
BCI %>% select(one_of(vector))
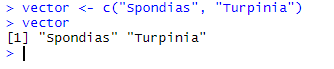
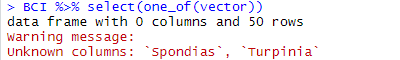
Note that you MUST store full names in the vector, not just parts of them. Here, we try with partial names:
vector <- c("Spondias", "Turpinia")
vector
And our previous code returns:
Finding the last variable with last_col()
last_col() finds the last column of the data frame.
BCI %>% select(last_col())
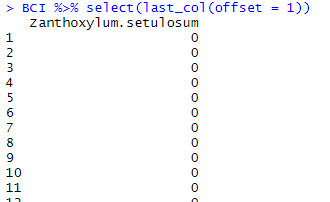
It may also retrieve the last column with an offset. If you indicate offset = 1, last_col() will retrieve the last but one column in the data frame:
BCI %>% select(last_col(offset = 1))