There are often situations where you need to update a data frame with additional cases, to merge different chronological versions of a data frame, to find out whether you have similar entries in two data frames,… in other words, situations where you need to identify and compare rows in two tables. Here we will review five useful functions that:
bind_rows(): place two tables on top of each other without further modificationsintersect(): return only the observations that are common to both tablessetdiff(): return only the observations which are specifically found in one table but not the other oneunion(): place two tables on top of each other while removing duplicatessetequal(): test whether two tables contain the exact same rows in any order
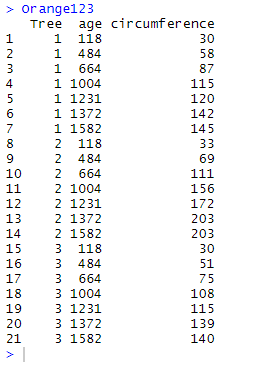
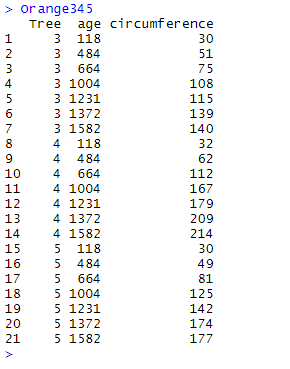
We will practically illustrate the use and purpose of the functions using Orange123 and Orange345 which are two complementary fragments of the data frame Orange. Note that the two fragments overlap (rows for which Tree equals 3). These fragments are generated using the following codes:
Orange123 <- Orange %>% filter(Tree == "1" | Tree == "2" | Tree == "3") Orange345 <- Orange %>% filter(Tree == "3" | Tree == "4" | Tree == "5")
Place two tables on top of each other without further modifications with bind_rows()
bind_rows() takes the content of two (or more) data frames and puts them under each other. The order of the data frames in the function defines the order of the rows in the output table. Note that NA values may be placed wherever bind_rows() does not find a value for the variable/observation; this occurs for example when binding data frames with unequal number of- or non matching variables. Also, bind_rows() does not check whether the output table has generated duplicates. Thus, if the original tables contain identical rows, the output table will contain several copies of these rows.
bind_rows(Orange123, Orange345)
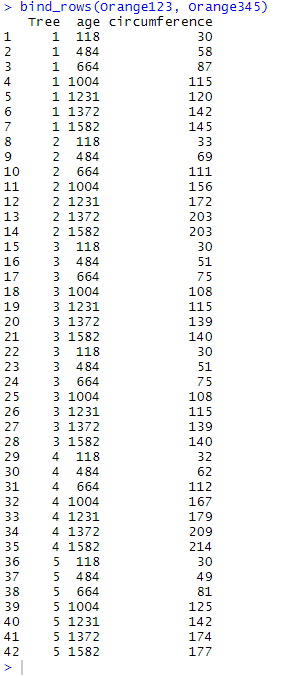
Here, Orange345 has been placed directly under Orange123. As you may see the rows where Tree equals 3 are duplicated since there was a copy of each of them in both Orange123 and Orange345.
Return only the observations that are common to both tables with intersect()
intersect() checks whether there are matching observations in the given data frames, and then returns only (one copy of) these observations in the output table.
intersect(Orange123, Orange345)
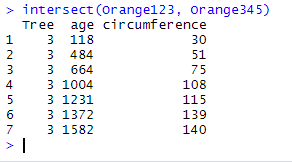
The function has found that there are 7 matching rows in Orange123 and Orange345, those where Tree equals 3, and has return a single copy of each.
Return only the observations which are specifically found in one table but not the other one with setdiff()
setdiff() compares the contents of two data frames to find out which observations are unique to the first named data frame, and then returns these observations in the output table. setdiff() will discard all observations which are common to both data frames, and will not consider any of the observations which are only found in the second data frame.
setdiff(Orange123, Orange345)
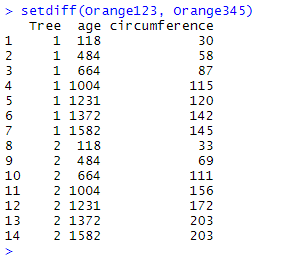
Here, setdiff() has only retained the observations for which Tree equals 1 or 2 since they are unique to Orange123, the first data frame named in the parentheses of the function.
Let’s see what happens when Orange345 is the first data frame:
setdiff(Orange345, Orange123)
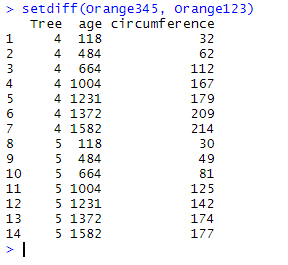
Here, setdiff() has only retained the observations for which Tree equals 4 or 5 since they are unique to Orange345.
Place two tables on top of each other while removing duplicates with union()
union() takes the contents of two (or more) data frames and puts them under each other, but only after having taken away the duplicates that arise from merging overlapping data frames.
union(Orange123, Orange345)
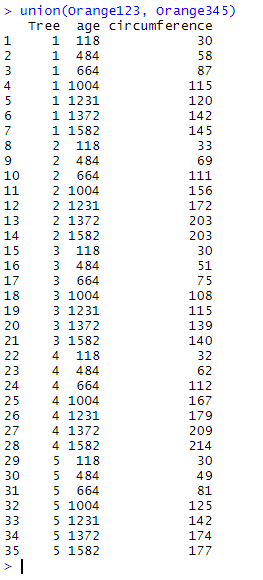
Here, Orange345 has been added under Orange123, but only one copy of the observations which are common to both data frames has been kept.
Test whether two tables contain the exact same rows in any order with setequal()
setequal() compares the contents of two data frames and tells whether their observations are identical or not by returning TRUE or FALSE. The order of observations within the data frames does not matter when comparing with setequal().
setequal(Orange123, Orange345)
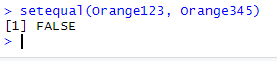
As expected, setequal() returns FALSE when comparing the contents of Orange123 and Orange345.
In the following example, we compare the original data frame Orange to the result of the “union” of Orange123 and Orange345 (which are two fragments of Orange) with union().
setequal(Orange, union(Orange123, Orange345))
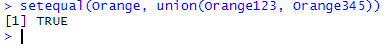
setequal() recognizes that the output of union(Orange123, Orange345) contains all the original observations found in Orange, nothing more, nothing less. It thus returns TRUE.
In this last example, we compare the original data frame Orange to the result of the concatenation of Orange123 and Orange345 with bind_rows().
setequal(Orange, bind_rows(Orange123, Orange345))
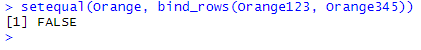
setequal() recognizes that the output of bind_rows(Orange123, Orange345) contains all the original observations found in Orange, but also finds duplicates that were not present originally. It thus returns FALSE.